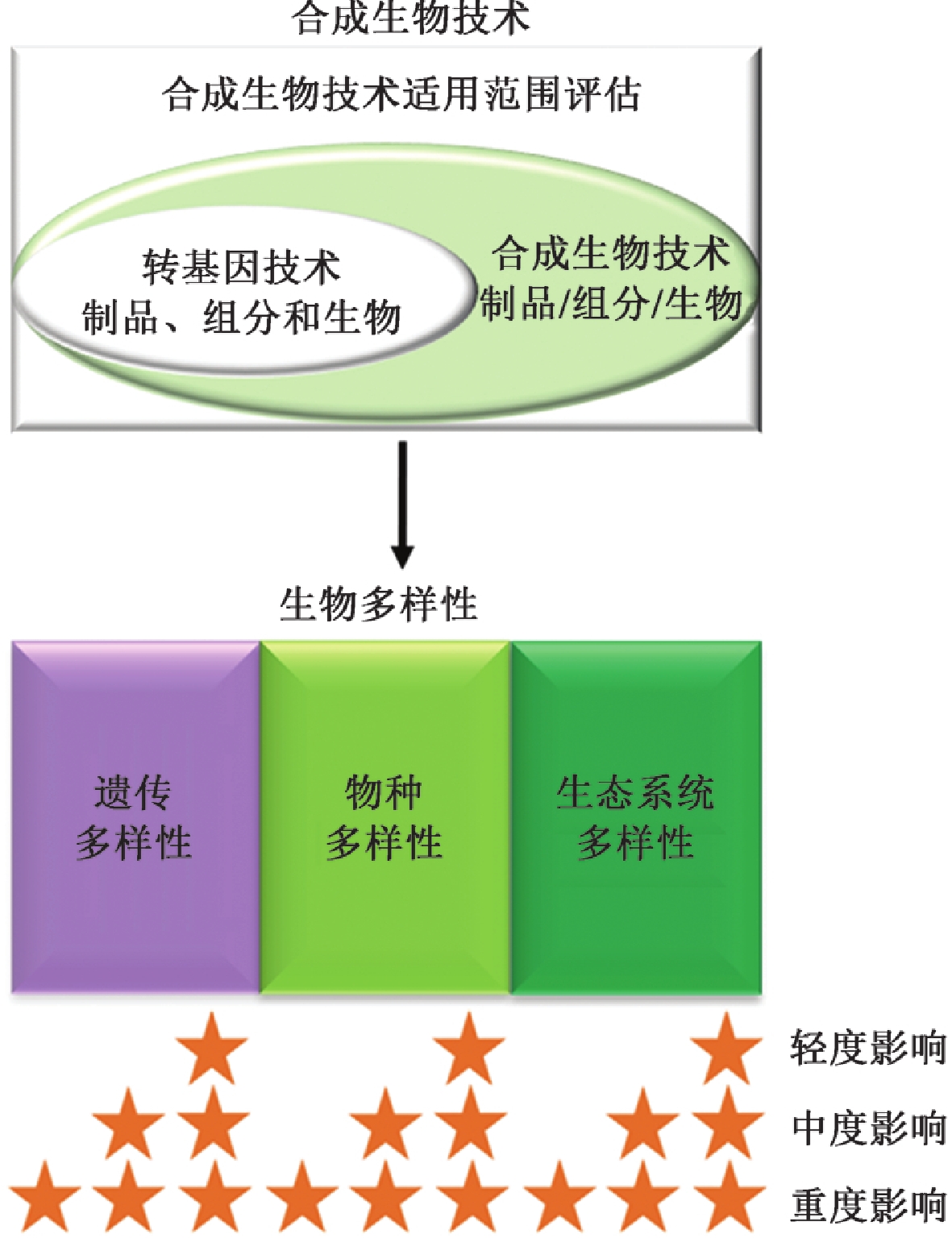
| [1] |
常江.《生物多样性公约》中合成生物学的谈判进程及对我国履约的启示[J]. 环境保护,2018,46(23):28-31.CHANG J. The negotiation process of synthetic in convention on biological diversity and its implications to China[J]. EnvironmentalProtection,2018,46(23):28-31.
|
| [2] |
宋健.保护生物多样性是全人类的共同责任[J]. 浙江林业,1994(4):1.
|
| [3] |
叶晓婷.我国是生物多样性最丰富12国之一 五大祸端威胁中国生物安全[J]. 环境与生活,2013(11):26-30.
|
| [4] |
冯晓娟, 米湘成, 肖治术, 等.中国生物多样性监测与研究网络建设及进展[J]. 中国科学院院刊,2019,34(12):1389-1398.FENG X J, MI X C, XIAO Z S, et al. Overview of Chinese biodiversity observation network (Sino BON)[J]. Bulletin of Chinese Academy of Sciences,2019,34(12):1389-1398.
|
| [5] |
李爱农, 尹高飞, 张正健, 等.基于站点的生物多样性星空地一体化遥感监测[J]. 生物多样性,2018,26(8):819-827. doi: 10.17520/biods.2018052LI A N, YIN G F, ZHANG Z J, et al. Space-air-field integrated biodiversity monitoring based on experimental station[J]. Biodiversity Science,2018,26(8):819-827. doi: 10.17520/biods.2018052
|
| [6] |
朱媛君, 山丹, 张晓, 等.揭示群落结构及其环境响应的联合物种分布模型的研究进展[J]. 应用生态学报,2018,29(12):4217-4225.ZHU Y J, SHAN D, ZHANG X, et al. Advances in joint species distribution models to reveal community structure and its environmental response[J]. Chinese Journal of Applied Ecology,2018,29(12):4217-4225.
|
| [7] |
马克平, 朱敏, 纪力强, 等.中国生物多样性大数据平台建设[J]. 中国科学院院刊,2018,33(8):838-845.MA K P, ZHU M, JI L Q, et al. Establishing China infrastructure for big biodiversity data[J]. Bulletin of Chinese Academy of Sciences,2018,33(8):838-845.
|
| [8] |
National Academies of Sciences, Engineering, and Medicine. Biodefense in the age of synthetic biology[M/OL]. Washington DC: The National Academies Press, 2018. https://doi.org/10.17226/24890.
|
| [9] |
中华人民共和国生物安全法[A/OL]. (2020-10-18)[2021-06-01]. http://www.xinhuanet.com/legal/2020-10/18/c_1126624481.htm.
|
| [10] |
BECKER J, WITTMANN C. Advanced biotechnology: metabolically engineered cells for the bio-based production of chemicals and fuels, materials, and health-care products[J]. Angewandte Chemie (International Edition in English),2015,54(11):3328-3350. doi: 10.1002/anie.201409033
|
| [11] |
NETT R S, LAU W, SATTELY E S. Discovery and engineering of colchicine alkaloid biosynthesis[J]. Nature,2020,584(7819):148-153. doi: 10.1038/s41586-020-2546-8
|
| [12] |
HOLKENBRINK C, DING B J, WANG H L, et al. Production of moth sex pheromones for pest control by yeast fermentation[J]. Metabolic Engineering,2020,62:312-321. doi: 10.1016/j.ymben.2020.10.001
|
| [13] |
WANG D, WANG J H, SHI Y S, et al. Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform[J]. Metabolic Engineering,2020,61:131-140. doi: 10.1016/j.ymben.2020.05.007
|
| [14] |
LOSOI P S, SANTALA V P, SANTALA S M. Enhanced population control in a synthetic bacterial consortium by interconnected carbon cross-feeding[J]. ACS Synthetic Biology,2019,8(12):2642-2650. doi: 10.1021/acssynbio.9b00316
|
| [15] |
KIM J K, CHEN Y, HIRNING A J, et al. Long-range temporal coordination of gene expression in synthetic microbial consortia[J]. Nature Chemical Biology,2019,15(11):1102-1109. doi: 10.1038/s41589-019-0372-9
|
| [16] |
GUTIÉRREZ N, GARRIDO D. Species deletions from microbiome consortia reveal key metabolic interactions between gut microbes[J]. mSystems, 2019, 4(4). doi: 10.1128/msystems.00185-19.
|
| [17] |
SADEGHPOUR M, VELIZ-CUBA A, OROSZ G, et al. Bistability and oscillations in co-repressive synthetic microbial consortia[J]. Quantitative Biology,2017,5(1):55-66. doi: 10.1007/s40484-017-0100-y
|
| [18] |
GARCÍA-JIMÉNEZ B, GARCÍA J L, NOGALES J. FLYCOP: metabolic modeling-based analysis and engineering microbial communities[J]. Bioinformatics,2018,34(17):i954-i963. doi: 10.1093/bioinformatics/bty561
|
| [19] |
MA Q, BI Y H, WANG E X, et al. Integrated proteomic and metabolomic analysis of a reconstructed three-species microbial consortium for one-step fermentation of 2-keto-L-gulonic acid, the precursor of vitamin C[J]. Journal of Industrial Microbiology & Biotechnology,2019,46(1):21-31.
|
| [20] |
MACCHI M, FESTA S, NIETO E, et al. Design and evaluation of synthetic bacterial consortia for optimized phenanthrene degradation through the integration of genomics and shotgun proteomics[J]. Biotechnology Reports,2021,29:e00588. doi: 10.1016/j.btre.2021.e00588
|
| [21] |
FENG S S, GONG L Q, ZHANG Y K, et al. Bioaugmentation potential evaluation of a bacterial consortium composed of isolated Pseudomonas and Rhodococcus for degrading benzene, toluene and styrene in sludge and sewage[J]. Bioresource Technology,2021,320:124329. doi: 10.1016/j.biortech.2020.124329
|
| [22] |
HUANG J M, YANG X N, WU Q H, et al. Application of independent immobilization in benzo[a]pyrene biodegradation by synthetic microbial consortium[J]. Environmental Science and Pollution Research,2019,26(20):21052-21058. doi: 10.1007/s11356-019-05477-4
|
| [23] |
KONG X X, JIANG J L, QIAO B, et al. The biodegradation of cefuroxime, cefotaxime and cefpirome by the synthetic consortium with probiotic Bacillus clausii and investigation of their potential biodegradation pathways[J]. Science of the Total Environment,2019,651:271-280. doi: 10.1016/j.scitotenv.2018.09.187
|
| [24] |
del VALLE I, FULK E M, KALVAPALLE P, et al. Translating new synthetic biology advances for biosensing into the earth and environmental sciences[J]. Frontiers in Microbiology,2021,11:618373. doi: 10.3389/fmicb.2020.618373
|
| [25] |
VANARSDALE E, TSAO C Y, LIU Y, et al. Redox-based synthetic biology enables electrochemical detection of the herbicides dicamba and roundup via rewired Escherichia coli[J]. ACS Sensors,2019,4(5):1180-1184. doi: 10.1021/acssensors.9b00085
|
| [26] |
CELLO J, PAUL A V, WIMMER E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template[J]. Science,2002,297(5583):1016-1018. doi: 10.1126/science.1072266
|
| [27] |
GIBSON D G, GLASS J I, LARTIGUE C, et al. Creation of a bacterial cell controlled by a chemically synthesized genome[J]. Science,2010,329(5987):52-56. doi: 10.1126/science.1190719
|
| [28] |
HUTCHISON C A, CHUANG R Y, NOSKOV V N, et al. Design and synthesis of a minimal bacterial genome[J]. Science,2016,351(6280):aad6253. doi: 10.1126/science.aad6253
|
| [29] |
WU Y, LI B Z, ZHAO M, et al. Bug mapping and fitness testing of chemically synthesized chromosome X[J]. Science,2017,355(6329):eaaf4706. doi: 10.1126/science.aaf4706
|
| [30] |
XIE Z X, LI B Z, MITCHELL L A, et al. "Perfect" designer chromosome V and behavior of a ring derivative[J]. Science,2017,355(6329):eaaf4704. doi: 10.1126/science.aaf4704
|
| [31] |
SHAO Y Y, LU N, WU Z F, et al. Creating a functional single-chromosome yeast[J]. Nature,2018,560(7718):331-335. doi: 10.1038/s41586-018-0382-x
|
| [32] |
BOEKE J D, CHURCH G, HESSEL A, et al. The genome project-write[J]. Science,2016,353(6295):126-127. doi: 10.1126/science.aaf6850
|
| [33] |
LEDFORD H, CALLAWAY E. Pioneers of revolutionary CRISPR gene editing win chemistry Nobel[J]. Nature,2020,586(7829):346-347. doi: 10.1038/d41586-020-02765-9
|
| [34] |
WANG D, ZHANG F, GAO G P. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors[J]. Cell,2020,181(1):136-150. doi: 10.1016/j.cell.2020.03.023
|
| [35] |
STORCH G A. CRISPR tool scales up to interrogate a huge line-up of viral suspects[J]. Nature,2020,582(7811):188-189. doi: 10.1038/d41586-020-01447-w
|
| [36] |
GAO H R, GADLAGE M J, LAFITTE H R, et al. Superior field performance of waxy corn engineered using CRISPR-Cas9[J]. Nature Biotechnology,2020,38(5):579-581. doi: 10.1038/s41587-020-0444-0
|
| [37] |
CHOI S Y, WOO H M. CRISPRi-dCas12a: a dCas12a-mediated CRISPR interference for repression of multiple genes and metabolic engineering in cyanobacteria[J]. ACS Synthetic Biology,2020,9(9):2351-2361. doi: 10.1021/acssynbio.0c00091
|
| [38] |
ABDEL-MAWGOUD A M, STEPHANOPOULOS G. Improving CRISPR/Cas9-mediated genome editing efficiency in Yarrowia lipolytica using direct tRNA-sgRNA fusions[J]. Metabolic Engineering,2020,62:106-115. doi: 10.1016/j.ymben.2020.07.008
|
| [39] |
SMITH C J, CASTANON O, SAID K, et al. Enabling large-scale genome editing at repetitive elements by reducing DNA nicking[J]. Nucleic Acids Research,2020,48(9):5183-5195. doi: 10.1093/nar/gkaa239
|
| [40] |
ZHAO D D, LI J, LI S W, et al. Glycosylase base editors enable C-to-A and C-to-G base changes[J]. Nature Biotechnology,2021,39(1):35-40. doi: 10.1038/s41587-020-0592-2
|
| [41] |
KLEINSTIVER B P, PATTANAYAK V, PREW M S, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects[J]. Nature,2016,529(7587):490-495. doi: 10.1038/nature16526
|
| [42] |
CHEN J S, DAGDAS Y S, KLEINSTIVER B P, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy[J]. Nature,2017,550(7676):407-410. doi: 10.1038/nature24268
|
| [43] |
SINKINS S P, GOULD F. Gene drive systems for insect disease vectors[J]. Nature Reviews Genetics,2006,7(6):427-435. doi: 10.1038/nrg1870
|
| [44] |
JAMES A A. Gene drive systems in mosquitoes: rules of the road[J]. Trends in Parasitology,2005,21(2):64-67. doi: 10.1016/j.pt.2004.11.004
|
| [45] |
KYROU K, HAMMOND A M, GALIZI R, et al. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes[J]. Nature Biotechnology,2018,36(11):1062-1066. doi: 10.1038/nbt.4245
|
| [46] |
PROWSE T A, ADIKUSUMA F, CASSEY P, et al. A Y-chromosome shredding gene drive for controlling pest vertebrate populations[J]. Elife,2019,8:e41873. doi: 10.7554/eLife.41873
|
| [47] |
CONKLIN B R. On the road to a gene drive in mammals[J]. Nature,2019,566(7742):43-45. doi: 10.1038/d41586-019-00185-y
|
| [48] |
NEVE P. Gene drive systems: do they have a place in agricultural weed management[J]. Pest Management Science,2018,74(12):2671-2679. doi: 10.1002/ps.5137
|
| [49] |
ECKHOFF P A, WENGER E A, GODFRAY H C J, et al. Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics[J]. Proceedings of the National Academy of Sciences of the United States of America,2017,114(2):E255-E264. doi: 10.1073/pnas.1611064114
|
| [50] |
BEAGHTON A, BEAGHTON P J, BURT A. Vector control with driving Y chromosomes: modelling the evolution of resistance[J]. Malaria Journal,2017,16(1):286. doi: 10.1186/s12936-017-1932-7
|
| [51] |
COLLINS J P. Gene drives in our future: challenges of and opportunities for using a self-sustaining technology in pest and vector management[J]. BMC Proceedings, 2018, 12(Suppl 8): 9.
|
| [52] |
JAMES S, COLLINS F H, WELKHOFF P A, et al. Pathway to deployment of gene drive mosquitoes as a potential biocontrol tool for elimination of malaria in sub-Saharan Africa: recommendations of a scientific working group[J]. American Journal of Tropical Medicine and Hygiene, 2018, 98(Suppl 6): 1-49.
|
| [53] |
钱万强, 江海燕, 朱庆平, 等.国内外合成生物学资助体系及产业投入分析[J]. 中国基础科学,2014,16(1):47-50. doi: 10.3969/j.issn.1009-2412.2014.01.008QIAN W Q, JIANG H Y, ZHU Q P, et al. Analysis of the funding systems and industry investment of synthetic biology in China and main developed countries[J]. China Basic Science,2014,16(1):47-50. doi: 10.3969/j.issn.1009-2412.2014.01.008
|
| [54] |
刘发鹏.合成生物学: 一种两用技术的机遇和挑战[J]. 科学中国人,2018(17):72-73.
|
| [55] |
YANG J G, XIE X Q, XIANG N, et al. Polyprotein strategy for stoichiometric assembly of nitrogen fixation components for synthetic biology[J]. Proceedings of the National Academy of Sciences of the United States of America,2018,115(36):8509-8517. doi: 10.1073/pnas.1804992115
|
| [56] |
WU M R, JUSIAK B, LU T K. Engineering advanced cancer therapies with synthetic biology[J]. Nature Reviews Cancer,2019,19(4):187-195.
|
| [57] |
MILLER T E, BENEYTON T, SCHWANDER T, et al. Light-powered CO2 fixation in a chloroplast mimic with natural and synthetic parts[J]. Science,2020,368(6491):649-654. doi: 10.1126/science.aaz6802
|
| [58] |
BOLES K S, KANNAN K, GILL J, et al. Digital-to-biological converter for on-demand production of biologics[J]. Nature Biotechnology,2017,35(7):672-675. doi: 10.1038/nbt.3859
|
| [59] |
MEHR S H M, CRAVEN M, LEONOV A I, et al. A universal system for digitization and automatic execution of the chemical synthesis literature[J]. Science,2020,370(6512):101-108. doi: 10.1126/science.abc2986
|
| [60] |
关正君, 裴蕾, 马库斯·施密特, 等.合成生物学生物安全风险评价与管理[J]. 生物多样性,2012,20(2):138-150.GUAN Z J, PEI L, SCHMIDT M, et al. Assessment and management of biosafety in synthetic biology[J]. Biodiversity Science,2012,20(2):138-150.
|
| [61] |
TANG T C, THAM E, LIU X Y, et al. Hydrogel-based biocontainment of bacteria for continuous sensing and computation[J]. Nature Chemical Biology,2021,17(6):724-731. doi: 10.1038/s41589-021-00779-6
|
| [62] |
YOO J I, SEPPÄLÄS O, MALLEY M A. Engineered fluoride sensitivity enables biocontainment and selection of genetically-modified yeasts[J]. Nature Communications,2020,11:5459. doi: 10.1038/s41467-020-19271-1
|
| [63] |
MASELKO M, HEINSCH S C, CHACÓN J M, et al. Engineering species-like barriers to sexual reproduction[J]. Nature Communications,2017,8(1):883. doi: 10.1038/s41467-017-01007-3
|
| [64] |
YOUNG R E B, PURTON S. Codon reassignment to facilitate genetic engineering and biocontainment in the chloroplast of Chlamydomonas reinhardtii[J]. Plant Biotechnology Journal,2016,14(5):1251-1260. doi: 10.1111/pbi.12490
|
| [65] |
CAI Y Z, AGMON N, CHOI W J, et al. Intrinsic biocontainment: multiplex genome safeguards combine transcriptional and recombinational control of essential yeast genes[J]. Proceedings of the National Academy of Sciences of the United States of America,2015,112(6):1803-1808. ⊗ doi: 10.1073/pnas.1424704112
|




 下载:
下载: